Complementary Filters Shaping Using \(\mathcal{H}_\infty\) Synthesis - Tikz Figures
Table of Contents
- 1. Fig 1: Sensor Fusion Architecture
- 2. Fig 2: Sensor fusion architecture with sensor dynamics uncertainty
- 3. Fig 3: Uncertainty set of the super sensor dynamics
- 4. Fig 4: Architecture used for \(\mathcal{H}_\infty\) synthesis of complementary filters
- 5. Fig 5: Magnitude of a weighting function generated using the proposed formula
- 6. Fig 6: Frequency response of the weighting functions and complementary filters obtained using \(\mathcal{H}_\infty\) synthesis
- 7. Fig 7: Architecture for \(\mathcal{H}_\infty\) synthesis of three complementary filters
- 8. Fig 8: Frequency response of the weighting functions and three complementary filters obtained using \(\mathcal{H}_\infty\) synthesis
- 9. Fig 9: Specifications and weighting functions magnitude used for \(\mathcal{H}_\infty\) synthesis
- 10. Fig 10: Comparison of the FIR filters (solid) with the filters obtained with \(\mathcal{H}_\infty\) synthesis (dashed)
Configuration file is accessible here.
1 Fig 1: Sensor Fusion Architecture
\begin{tikzpicture} \node[branch] (x) at (0, 0); \node[block, above right=0.5 and 0.5 of x](G1){$G_1(s)$}; \node[block, below right=0.5 and 0.5 of x](G2){$G_2(s)$}; \node[addb, right=0.8 of G1](add1){}; \node[addb, right=0.8 of G2](add2){}; \node[block, right=0.8 of add1](H1){$H_1(s)$}; \node[block, right=0.8 of add2](H2){$H_2(s)$}; \node[addb, right=5 of x](add){}; \draw[] ($(x)+(-0.7, 0)$) node[above right]{$x$} -- (x.center); \draw[->] (x.center) |- (G1.west); \draw[->] (x.center) |- (G2.west); \draw[->] (G1.east) -- (add1.west); \draw[->] (G2.east) -- (add2.west); \draw[<-] (add1.north) -- ++(0, 0.8)node[below right](n1){$n_1$}; \draw[<-] (add2.north) -- ++(0, 0.8)node[below right](n2){$n_2$}; \draw[->] (add1.east) -- (H1.west); \draw[->] (add2.east) -- (H2.west); \draw[->] (H1) -| (add.north); \draw[->] (H2) -| (add.south); \draw[->] (add.east) -- ++(0.7, 0) node[above left]{$\hat{x}$}; \begin{scope}[on background layer] \node[fit={($(G2.south-|x)+(-0.2, -0.3)$) ($(n1.north east-|add.east)+(0.2, 0.3)$)}, fill=black!10!white, draw, dashed, inner sep=0pt] (supersensor) {}; \node[below left] at (supersensor.north east) {Super Sensor}; \node[fit={($(G1.south west)+(-0.3, -0.1)$) ($(n1.north east)+(0.0, 0.1)$)}, fill=black!20!white, draw, dashed, inner sep=0pt] (sensor1) {}; \node[below right] at (sensor1.north west) {Sensor 1}; \node[fit={($(G2.south west)+(-0.3, -0.1)$) ($(n2.north east)+(0.0, 0.1)$)}, fill=black!20!white, draw, dashed, inner sep=0pt] (sensor2) {}; \node[below right] at (sensor2.north west) {Sensor 2}; \end{scope} \end{tikzpicture}
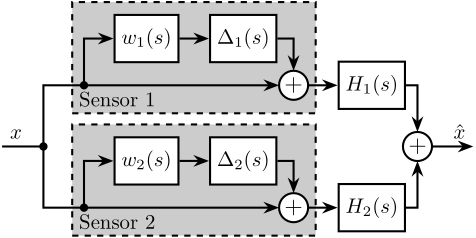
2 Fig 2: Sensor fusion architecture with sensor dynamics uncertainty
\begin{tikzpicture} \node[branch] (x) at (0, 0); \node[addb, above right=0.8 and 4 of x](add1){}; \node[addb, below right=0.8 and 4 of x](add2){}; \node[block, above left=0.2 and 0.1 of add1](delta1){$\Delta_1(s)$}; \node[block, above left=0.2 and 0.1 of add2](delta2){$\Delta_2(s)$}; \node[block, left=0.5 of delta1](W1){$w_1(s)$}; \node[block, left=0.5 of delta2](W2){$w_2(s)$}; \node[block, right=0.5 of add1](H1){$H_1(s)$}; \node[block, right=0.5 of add2](H2){$H_2(s)$}; \node[addb, right=6 of x](add){}; \draw[] ($(x)+(-0.7, 0)$) node[above right]{$x$} -- (x.center); \draw[->] (x.center) |- (add1.west); \draw[->] (x.center) |- (add2.west); \draw[->] ($(add1-|W1.west)+(-0.5, 0)$)node[branch](S1){} |- (W1.west); \draw[->] ($(add2-|W2.west)+(-0.5, 0)$)node[branch](S1){} |- (W2.west); \draw[->] (W1.east) -- (delta1.west); \draw[->] (W2.east) -- (delta2.west); \draw[->] (delta1.east) -| (add1.north); \draw[->] (delta2.east) -| (add2.north); \draw[->] (add1.east) -- (H1.west); \draw[->] (add2.east) -- (H2.west); \draw[->] (H1.east) -| (add.north); \draw[->] (H2.east) -| (add.south); \draw[->] (add.east) -- ++(0.7, 0) node[above left]{$\hat{x}$}; \begin{scope}[on background layer] \node[block, fit={($(W1.north-|S1)+(-0.2, 0.2)$) ($(add1.south east)+(0.2, -0.3)$)}, fill=black!20!white, dashed, inner sep=0pt] (sensor1) {}; \node[above right] at (sensor1.south west) {Sensor 1}; \node[block, fit={($(W2.north-|S1)+(-0.2, 0.2)$) ($(add2.south east)+(0.2, -0.3)$)}, fill=black!20!white, dashed, inner sep=0pt] (sensor2) {}; \node[above right] at (sensor2.south west) {Sensor 2}; \end{scope} \end{tikzpicture}
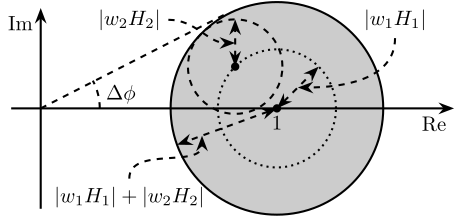
3 Fig 3: Uncertainty set of the super sensor dynamics
\begin{tikzpicture} \begin{scope}[shift={(4, 0)}] % Uncertainty Circle \node[draw, circle, fill=black!20!white, minimum size=3.6cm] (c) at (0, 0) {}; \path[draw, dotted] (0, 0) circle [radius=1.0]; \path[draw, dashed] (135:1.0) circle [radius=0.8]; % Center of Circle \node[below] at (0, 0){$1$}; \draw[<->, dashed] (0, 0) node[branch]{} -- coordinate[midway](r1) ++(45:1.0); \draw[<->, dashed] (135:1.0)node[branch]{} -- coordinate[midway](r2) ++(90:0.8); \node[] (l1) at (2, 1.5) {$|w_1 H_1|$}; \draw[->, dashed, out=-90, in=0] (l1.south) to (r1); \node[] (l2) at (-2.5, 1.5) {$|w_2 H_2|$}; \draw[->, dashed, out=0, in=-180] (l2.east) to (r2); \draw[<->, dashed] (0, 0) -- coordinate[near end](r3) ++(200:1.8); \node[] (l3) at (-2.5, -1.5) {$|w_1 H_1| + |w_2 H_2|$}; \draw[->, dashed, out=90, in=-90] (l3.north) to (r3); \end{scope} % Real and Imaginary Axis \draw[->] (-0.5, 0) -- (7.0, 0) node[below left]{Re}; \draw[->] (0, -1.7) -- (0, 1.7) node[below left]{Im}; \draw[dashed] (0, 0) -- (tangent cs:node=c,point={(0, 0)},solution=2); \draw[dashed] (1, 0) arc (0:28:1) node[midway, right]{$\Delta \phi$}; \end{tikzpicture}
4 Fig 4: Architecture used for \(\mathcal{H}_\infty\) synthesis of complementary filters
\begin{tikzpicture} \node[block={4.0cm}{2.5cm}, fill=black!20!white, dashed] (P) {}; \node[above] at (P.north) {$P(s)$}; \coordinate[] (inputw) at ($(P.south west)!0.75!(P.north west) + (-0.7, 0)$); \coordinate[] (inputu) at ($(P.south west)!0.35!(P.north west) + (-0.7, 0)$); \coordinate[] (output1) at ($(P.south east)!0.75!(P.north east) + ( 0.7, 0)$); \coordinate[] (output2) at ($(P.south east)!0.35!(P.north east) + ( 0.7, 0)$); \coordinate[] (outputv) at ($(P.south east)!0.1!(P.north east) + ( 0.7, 0)$); \node[block, left=1.4 of output1] (W1){$W_1(s)$}; \node[block, left=1.4 of output2] (W2){$W_2(s)$}; \node[addb={+}{}{}{}{-}, left=of W1] (sub) {}; \node[block, below=0.3 of P] (H2) {$H_2(s)$}; \draw[->] (inputw) node[above right]{$w$} -- (sub.west); \draw[->] (H2.west) -| ($(inputu)+(0.35, 0)$) node[above]{$u$} -- (W2.west); \draw[->] (inputu-|sub) node[branch]{} -- (sub.south); \draw[->] (sub.east) -- (W1.west); \draw[->] ($(sub.west)+(-0.6, 0)$) node[branch]{} |- ($(outputv)+(-0.35, 0)$) node[above]{$v$} |- (H2.east); \draw[->] (W1.east) -- (output1)node[above left]{$z_1$}; \draw[->] (W2.east) -- (output2)node[above left]{$z_2$}; \end{tikzpicture}
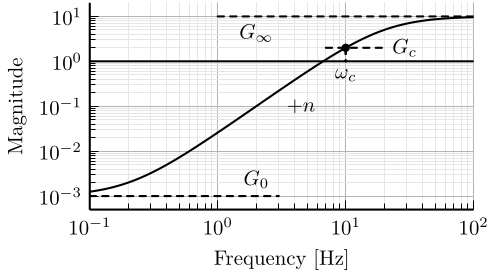
5 Fig 5: Magnitude of a weighting function generated using the proposed formula
\setlength\fwidth{6.5cm} \setlength\fheight{3.5cm} \begin{tikzpicture} \begin{axis}[% width=1.0\fwidth, height=1.0\fheight, at={(0.0\fwidth, 0.0\fheight)}, scale only axis, xmode=log, xmin=0.1, xmax=100, xtick={0.1,1,10, 100}, xminorticks=true, ymode=log, ymin=0.0005, ymax=20, ytick={0.001, 0.01, 0.1, 1, 10}, yminorticks=true, ylabel={Magnitude}, xlabel={Frequency [Hz]}, xminorgrids, yminorgrids, ] \addplot [color=black, line width=1.5pt, forget plot] table [x=freqs, y=ampl, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matweight_formula.csv}; \addplot [color=black, dashed, line width=1.5pt] table[row sep=crcr]{% 1 10\\ 100 10\\ }; \addplot [color=black, dashed, line width=1.5pt] table[row sep=crcr]{% 0.1 0.001\\ 3 0.001\\ }; \addplot [color=black, line width=1.5pt] table[row sep=crcr]{% 0.1 1\\ 100 1\\ }; \addplot [color=black, dashed, line width=1.5pt] table[row sep=crcr]{% 10 2\\ 10 1\\ }; \node[below] at (2, 10) {$G_\infty$}; \node[above] at (2, 0.001) {$G_0$}; \node[branch] at (10, 2){}; \draw[dashed, line cap=round] (7, 2) -- (20, 2) node[right]{$G_c$}; \draw[dashed, line cap=round] (10, 2) -- (10, 1) node[below]{$\omega_c$}; \node[right] at (3, 0.1) {$+n$}; \end{axis} \end{tikzpicture}
6 Fig 6: Frequency response of the weighting functions and complementary filters obtained using \(\mathcal{H}_\infty\) synthesis
\setlength\fwidth{6.5cm} \setlength\fheight{6cm} \begin{tikzpicture} \begin{axis}[% width=1.0\fwidth, height=0.5\fheight, at={(0.0\fwidth, 0.47\fheight)}, scale only axis, xmode=log, xmin=0.1, xmax=1000, xtick={0.1, 1, 10, 100, 1000}, xticklabels={{}}, xminorticks=true, ymode=log, ymin=0.0005, ymax=20, ytick={0.001, 0.01, 0.1, 1, 10}, yminorticks=true, ylabel={Magnitude}, xminorgrids, yminorgrids, ] \addplot [color=mycolor1, line width=1.5pt, forget plot] table [x=freqs, y=H1, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_filters_results.csv}; \addplot [color=mycolor2, line width=1.5pt, forget plot] table [x=freqs, y=H2, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_filters_results.csv}; \addplot [color=mycolor1, dashed, line width=1.5pt, forget plot] table [x=freqs, y=W1, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_weights.csv}; \addplot [color=mycolor2, dashed, line width=1.5pt, forget plot] table [x=freqs, y=W2, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_weights.csv}; \end{axis} \begin{axis}[% width=1.0\fwidth, height=0.45\fheight, at={(0.0\fwidth, 0.0\fheight)}, scale only axis, xmode=log, xmin=0.1, xmax=1000, xtick={0.1, 1, 10, 100, 1000}, xminorticks=true, xlabel={Frequency [Hz]}, ymin=-200, ymax=200, ytick={-180, -90, 0, 90, 180}, ylabel={Phase [deg]}, xminorgrids, legend style={at={(1,1.1)}, outer sep=2pt , anchor=north east, legend cell align=left, align=left, draw=black, nodes={scale=0.7, transform shape}}, ] \addlegendimage{color=mycolor1, dashed, line width=1.5pt} \addlegendentry{$W_1^{-1}$}; \addlegendimage{color=mycolor2, dashed, line width=1.5pt} \addlegendentry{$W_2^{-1}$}; \addplot [color=mycolor1, line width=1.5pt] table [x=freqs, y=H1p, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_filters_results.csv}; \addlegendentry{$H_1$}; \addplot [color=mycolor2, line width=1.5pt] table [x=freqs, y=H2p, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_filters_results.csv}; \addlegendentry{$H_2$}; \end{axis} \end{tikzpicture}
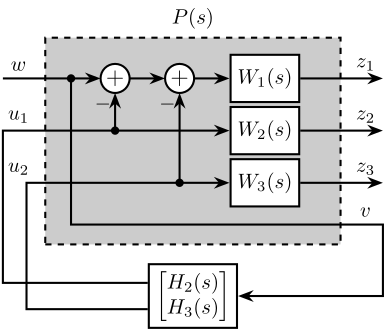
7 Fig 7: Architecture for \(\mathcal{H}_\infty\) synthesis of three complementary filters
\begin{tikzpicture} \node[block={5.0cm}{3.5cm}, fill=black!20!white, dashed] (P) {}; \node[above] at (P.north) {$P(s)$}; \coordinate[] (inputw) at ($(P.south west)!0.8!(P.north west) + (-0.7, 0)$); \coordinate[] (inputu) at ($(P.south west)!0.4!(P.north west) + (-0.7, 0)$); \coordinate[] (output1) at ($(P.south east)!0.8!(P.north east) + (0.7, 0)$); \coordinate[] (output2) at ($(P.south east)!0.55!(P.north east) + (0.7, 0)$); \coordinate[] (output3) at ($(P.south east)!0.3!(P.north east) + (0.7, 0)$); \coordinate[] (outputv) at ($(P.south east)!0.1!(P.north east) + (0.7, 0)$); \node[block, left=1.4 of output1] (W1){$W_1(s)$}; \node[block, left=1.4 of output2] (W2){$W_2(s)$}; \node[block, left=1.4 of output3] (W3){$W_3(s)$}; \node[addb={+}{}{}{}{-}, left=of W1] (sub1) {}; \node[addb={+}{}{}{}{-}, left=of sub1] (sub2) {}; \node[block, below=0.3 of P] (H) {$\begin{bmatrix}H_2(s) \\ H_3(s)\end{bmatrix}$}; \draw[->] (inputw) node[above right](w){$w$} -- (sub2.west); \draw[->] (W3-|sub1)node[branch]{} -- (sub1.south); \draw[->] (W2-|sub2)node[branch]{} -- (sub2.south); \draw[->] ($(sub2.west)+(-0.5, 0)$) node[branch]{} |- (outputv) |- (H.east); \draw[->] ($(H.south west)!0.7!(H.north west)$) -| (inputu|-W2) -- (W2.west); \draw[->] ($(H.south west)!0.3!(H.north west)$) -| ($(inputu|-W3)+(0.4, 0)$) -- (W3.west); \draw[->] (sub2.east) -- (sub1.west); \draw[->] (sub1.east) -- (W1.west); \draw[->] (W1.east) -- (output1)node[above left](z){$z_1$}; \draw[->] (W2.east) -- (output2)node[above left]{$z_2$}; \draw[->] (W3.east) -- (output3)node[above left]{$z_3$}; \node[above] at (W2-|w){$u_1$}; \node[above] at (W3-|w){$u_2$}; \node[above] at (outputv-|z){$v$}; \end{tikzpicture}
8 Fig 8: Frequency response of the weighting functions and three complementary filters obtained using \(\mathcal{H}_\infty\) synthesis
\setlength\fwidth{6.5cm} \setlength\fheight{6cm} \begin{tikzpicture} \begin{axis}[% width=1.0\fwidth, height=0.55\fheight, at={(0.0\fwidth, 0.42\fheight)}, scale only axis, xmode=log, xmin=0.1, xmax=100, xticklabels={{}}, xminorticks=true, ymode=log, ymin=0.0005, ymax=20, ytick={0.001, 0.01, 0.1, 1, 10}, yminorticks=true, ylabel={Magnitude}, xminorgrids, yminorgrids, legend columns=2, legend style={ /tikz/column 2/.style={ column sep=5pt, }, at={(1,0)}, outer sep=2pt , anchor=south east, legend cell align=left, align=left, draw=black, nodes={scale=0.7, transform shape} }, ] \addplot [color=mycolor1, dashed, line width=1.5pt] table [x=freqs, y=W1, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_three_weights.csv}; \addlegendentry{${W_1}^{-1}$}; \addplot [color=mycolor1, line width=1.5pt] table [x=freqs, y=H1, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_three_results.csv}; \addlegendentry{$H_1$}; \addplot [color=mycolor2, dashed, line width=1.5pt] table [x=freqs, y=W2, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_three_weights.csv}; \addlegendentry{${W_2}^{-1}$}; \addplot [color=mycolor2, line width=1.5pt] table [x=freqs, y=H2, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_three_results.csv}; \addlegendentry{$H_2$}; \addplot [color=mycolor3, dashed, line width=1.5pt] table [x=freqs, y=W3, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_three_weights.csv}; \addlegendentry{${W_3}^{-1}$}; \addplot [color=mycolor3, line width=1.5pt] table [x=freqs, y=H3, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_three_results.csv}; \addlegendentry{$H_3$}; \end{axis} \begin{axis}[% width=1.0\fwidth, height=0.4\fheight, at={(0.0\fwidth, 0.0\fheight)}, scale only axis, xmode=log, xmin=0.1, xmax=100, xminorticks=true, xlabel={Frequency [Hz]}, ymin=-240, ymax=240, ytick={-180, -90, 0, 90, 180}, ylabel={Phase [deg]}, xminorgrids, ] \addplot [color=mycolor1, line width=1.5pt] table [x=freqs, y=H1p, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_three_results.csv}; \addplot [color=mycolor2, line width=1.5pt] table [x=freqs, y=H2p, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_three_results.csv}; \addplot [color=mycolor3, line width=1.5pt] table [x=freqs, y=H3p, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/mathinf_three_results.csv}; \end{axis} \end{tikzpicture}
9 Fig 9: Specifications and weighting functions magnitude used for \(\mathcal{H}_\infty\) synthesis
\setlength\fwidth{6.5cm} \setlength\fheight{3.2cm} \begin{tikzpicture} \begin{axis}[% width=1.0\fwidth, height=1.0\fheight, at={(0.0\fwidth, 0.0\fheight)}, scale only axis, separate axis lines, every outer x axis line/.append style={black}, every x tick label/.append style={font=\color{black}}, every x tick/.append style={black}, xmode=log, xmin=0.001, xmax=1, xminorticks=true, xlabel={Frequency [Hz]}, every outer y axis line/.append style={black}, every y tick label/.append style={font=\color{black}}, every y tick/.append style={black}, ymode=log, ymin=0.005, ymax=20, yminorticks=true, ylabel={Magnitude}, axis background/.style={fill=white}, xmajorgrids, xminorgrids, ymajorgrids, yminorgrids, legend style={at={(0,1)}, outer sep=2pt, anchor=north west, legend cell align=left, align=left, draw=black, nodes={scale=0.7, transform shape}} ] \addplot [color=mycolor1, line width=1.5pt] table [x=freqs, y=wHm, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matligo_weights.csv}; \addlegendentry{$|w_H|^{-1}$} \addplot [color=mycolor2, line width=1.5pt] table [x=freqs, y=wLm, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matligo_weights.csv}; \addlegendentry{$|w_L|^{-1}$} \addplot [color=black, dotted, line width=1.5pt] table[row sep=crcr]{% 0.0005 0.008\\ 0.008 0.008\\ }; \addlegendentry{Specifications} \addplot [color=black, dotted, line width=1.5pt, forget plot] table[row sep=crcr]{% 0.008 0.008\\ 0.04 1\\ }; \addplot [color=black, dotted, line width=1.5pt, forget plot] table[row sep=crcr]{% 0.04 3\\ 0.1 3\\ }; \addplot [color=black, dotted, line width=1.5pt] table[row sep=crcr]{% 0.1 0.045\\ 2 0.045\\ }; \end{axis} \end{tikzpicture}
10 Fig 10: Comparison of the FIR filters (solid) with the filters obtained with \(\mathcal{H}_\infty\) synthesis (dashed)
\setlength\fwidth{6.5cm} \setlength\fheight{6.8cm} \begin{tikzpicture} \begin{axis}[% width=1.0\fwidth, height=0.60\fheight, at={(0.0\fwidth, 0.32\fheight)}, scale only axis, xmode=log, xmin=0.001, xmax=1, xtick={0.001,0.01,0.1,1}, xticklabels={{}}, xminorticks=true, ymode=log, ymin=0.002, ymax=5, ytick={0.001, 0.01, 0.1, 1, 10}, yminorticks=true, ylabel={Magnitude}, xminorgrids, yminorgrids, legend style={at={(1,0)}, outer sep=2pt, anchor=south east, legend cell align=left, align=left, draw=black, nodes={scale=0.7, transform shape}} ] \addplot [color=mycolor1, line width=1.5pt] table [x=freqs, y=Hhm, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matcomp_ligo_hinf.csv}; \addlegendentry{$H_H(s)$ - $\mathcal{H}_\infty$} \addplot [color=mycolor1, dashed, line width=1.5pt] table [x=freqs, y=Hhm, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matcomp_ligo_fir.csv}; \addlegendentry{$H_H(s)$ - FIR} \addplot [color=mycolor2, line width=1.5pt] table [x=freqs, y=Hlm, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matcomp_ligo_hinf.csv}; \addlegendentry{$H_L(s)$ - $\mathcal{H}_\infty$} \addplot [color=mycolor2, dashed, line width=1.5pt] table [x=freqs, y=Hlm, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matcomp_ligo_fir.csv}; \addlegendentry{$H_L(s)$ - FIR} \end{axis} \begin{axis}[% width=1.0\fwidth, height=0.3\fheight, at={(0.0\fwidth, 0.0\fheight)}, scale only axis, xmode=log, xmin=0.001, xmax=1, xtick={0.001, 0.01, 0.1, 1}, xminorticks=true, xlabel={Frequency [Hz]}, ymin=-180, ymax=180, ytick={-180, -90, 0, 90, 180}, ylabel={Phase [deg]}, xminorgrids, ] \addplot [color=mycolor1, line width=1.5pt, forget plot] table [x=freqs, y=Hhp, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matcomp_ligo_hinf.csv}; \addplot [color=mycolor1, dashed, line width=1.5pt, forget plot] table [x=freqs, y=Hhp, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matcomp_ligo_fir.csv}; \addplot [color=mycolor2, line width=1.5pt, forget plot] table [x=freqs, y=Hlp, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matcomp_ligo_hinf.csv}; \addplot [color=mycolor2, dashed, line width=1.5pt, forget plot] table [x=freqs, y=Hlp, col sep=comma] {/home/thomas/Cloud/thesis/papers/dehaeze19_desig_compl_filte/matlab/matcomp_ligo_fir.csv}; \end{axis} \end{tikzpicture}